RESEARCH
Here you’ll find detailed information on our main research areas.
Follow the links and learn more about each one of them.
Lysophosphatidic acid (LPA) receptors
Lysophosphatidic acid (LPA) is a bioactive lipid that signals through six different G protein-coupled receptors (LPA1–6) to mediate unique and overlapping biological effects. Thus, LPA plays key regulatory roles in pain, inflammation, cancer, and aging, mainly mediated by LPA1, LPA2 and LPA3 receptors. Considering the lack of potent and selective agonists and antagonists with receptor subtype specificity, our research group has addressed the development of new ligands to elucidate the precise mechanisms of the role of LPA in pain and neuroinflammation, and to validate the therapeutic potential of the receptors involved. To date, we have identified LPA1 agonists for the treatment of neuropathic pain and LPA2 antagonists able to ameliorate spinal cord injury. We continue our efforts to develop new LPA ligands with improved biological and pharmacokinetic profiles that could provide new pharmacological strategies to treat neuroinflammation-related pathologies.
This project is carried out in collaboration with Prof. Rubèn López Vales (Universidad Autónoma de Barcelona) and Prof. Jerold Chun (Sanford Burnham Prebys Medical Discovery Institute, California, USA).

Nucleophosmin 1 (NPM1) protein
Acute myeloid leukemia (AML) is a disease caused by myeloid hematological cancer stem cells (CSCs) with a high mortality rate and a low 5-year survival rate (around 30%). The most common mutations found in AML occur in Flt3 and Npm1 genes. While FLT3 mutation has targeted therapies such as midostaurin (Rydapt®), no NPM1 inhibitor has yet been successfully translated into clinical practice.
Following a phenotypic drug discovery strategy using CSC models and a proprietary focused library of microbiota-inspired compounds, UCM-13369 was identified as a hit capable of inhibiting CSC growth, without cytotoxicity in non-tumor cells. Differential proteomic studies together with Western blot analysis, confocal microscopy and ITC experiments revealed that the activity of UCM-13369 was linked to NPM1 protein. The efficacy of UCM-13369 was confirmed ex vivo in blood samples from AML patients and in vivo in a mouse xenograft model, strongly supporting the therapeutic potential of the identified NPM1 inhibitor for the treatment of AML. Based on these results, we have undertaken a medicinal chemistry program around UCM-13369, aimed at fully establishing the preclinical potential of the optimized compounds with the ultimate goal of advancing the best candidate towards clinical Phase I.
This project is carried out in collaboration with Dr. Miguel Gallardo (CNIO and Hospital Doce de Octubre, Madrid) and Prof. Irene Díaz-Moreno (Universidad de Sevilla and cicCartuja CSIC, Seville).
New Antibiotics for Resistant Bacterial Infections
Antibacterial resistance is one of the health biggest threats in the 21st century and novel approaches for multidrug resistant (MDR) infections are urgently required. In this regard, we are working on new pharmacological strategies focused on the discovery of antimicrobial agents with new mechanisms of action and the development of more efficacious treatments based on targeted drug delivery.
In the search for new antibiotics, we have focused our efforts on the inhibition of the bacterial cell division process by targeting the key protein FtsZ. Allosteric binding of small molecules to FtsZ can impair its function and eventually inhibit bacterial division. We have developed the first fluorescent competitive binding assay to identify FtsZ allosteric ligands, which will enable the identification of more effective antibacterials.
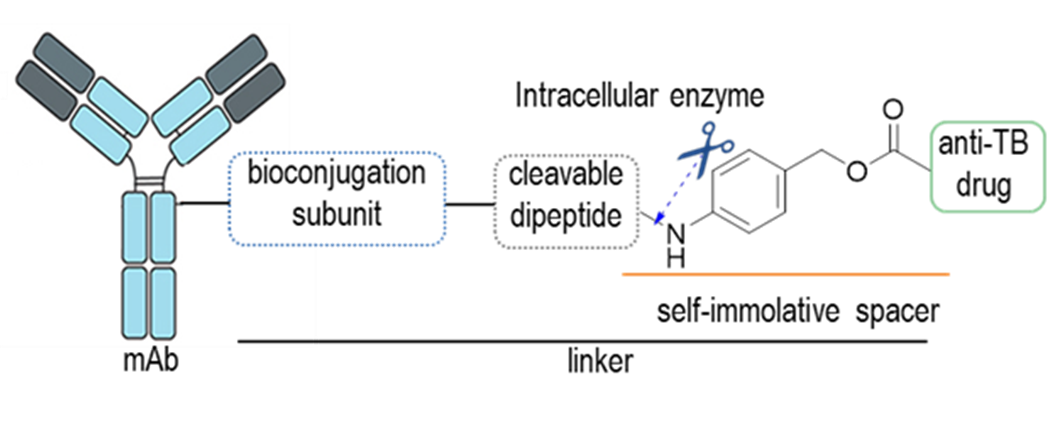
The global concern of MDR infections is particularly important in the tuberculosis (TB) field. The low efficacy of the current treatments for TB is mainly due to their poor capacity to reach the granuloma. In this context, our group is working on the development of antibody-antibiotic conjugates (AACs) that could specifically eliminate M. tuberculosis by directing the drug to the granuloma to eliminate these bacterial niches. The strategy encompasses the attachment of a monoclonal antibody (mAb), directed to M. tuberculosis or to antigens in the surface of the macrophages of mammalian cells, to an anti-TB drug that is selectively released inside the infected cell through a cleavable linker.
This project is carried out in collaboration with Dr. Rafael Prados (Universidad Autónoma de Madrid).
Progeria
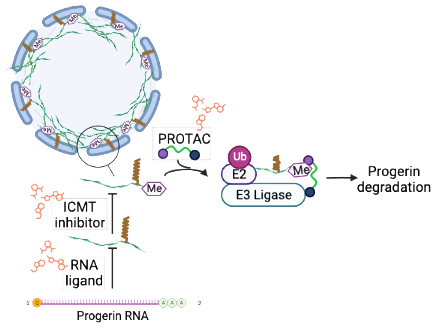
Hutchinson-Gilford Progeria Syndrome (HGPS or progeria) is a rare, fatal genetic condition characterized by an appearance of accelerated aging in children. Its name is derived from the Greek and means “prematurely old”. HGPS is caused by a mutation in the Lmna gene. This mutation produces progerin, a defective Lamin A protein. Progerin abnormally accumulates in the cell nucleus, thus affecting the majority of cellular processes and leading to the death of the patients at a median age of 15 years. It is believed that specific removal of progerin could lead to significant amelioration of the disease.
Within this aim, we are addressing the reduction of the progerin levels by three different strategies: (i) an indirect approach aimed at the inhibition of the enzyme isoprenylcysteine carboxylmethyltransferase (ICMT), (ii) the direct targeting of progerin by using proteolysis targeting chimeras (PROTACs) and (iii) the development of new RNA ligands.
This project is carried out in collaboration with Dr. Vicente Andrés (CNIC, Madrid) and Prof. M.D. Disney (The Scripps Research Institute, Florida, USA).
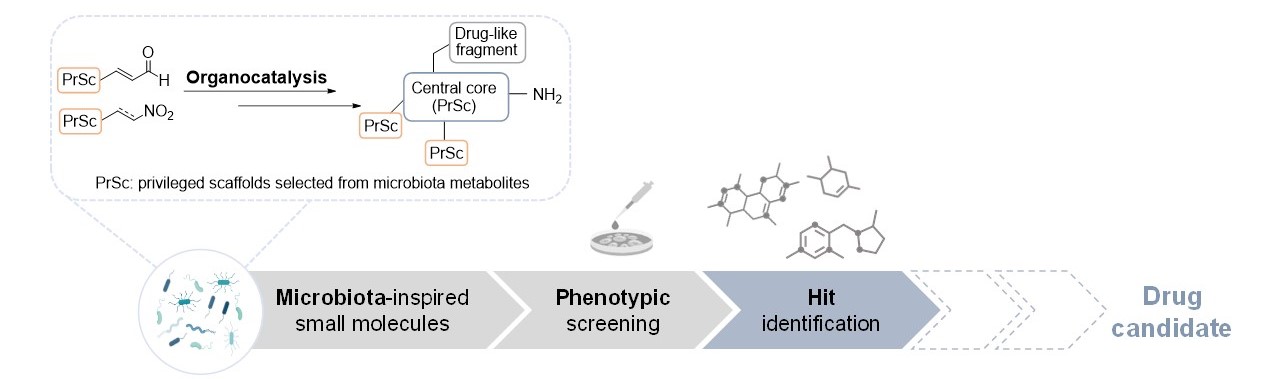
Phenotypic drug discovery using microbiota-inspired small molecules
Microbiota plays an important role in human health and disease, in part through the secretion of metabolites that can regulate host proteins triggering the modulation of metabolic, immune, and neuroendocrine pathways. Thus, metabolites produced by human microbiota can be considered drug-like specific modulators and include a diverse array of bioactive molecules. We propose that microbiota metabolites contain privileged structural scaffolds that represent an unexplored chemical space in the search for initial hits for drug discovery programs. Toward this end, we have analyzed the structures of identified metabolites to design a focused library of novel structurally diverse chemotypes based on privileged scaffolds present in selected metabolites. For the synthesis of the ~50 designed compounds we have applied a general strategy that relies on asymmetric aminocatalytic reactions to construct the central core in an efficient and stereocontrolled manner.
Our microbiota-inspired library has been explored in relevant cellular phenotypes following a phenotypic drug discovery strategy. In a cancer-stem-cell (CSC) model we identified a hit able of inhibiting CSC growth, without cytotoxicity in non-tumor cells. Further studies revealed that this compound acts by inhibiting NPM1, a protein involved in acute myeloid leukemia (AML). The compound has entered a medicinal chemistry program with the ultimate goal of obtaining a clinical candidate as a precision treatment for AML with mutated NPM1. In a second phenotypic screening of the microbiota-inspired compounds using a cellular model of senescence, we identified a hit that reduced senescence in human fibroblasts without toxicity in normal cells. Structurally related analogues were synthesized; selected compounds endowed with in vitro senolytic activity, no cytotoxicity, and appropriate ADME profile are currently under evaluation in animal models of pulmonary fibrosis and cancer to confirm their senotherapeutic efficacy in vivo and their potential for the treatment of age-related diseases, one of the most pressing biomedical challenges in our society. These results validate our phenotypic approach based on microbiota-inspired molecules as an innovative starting point for the identification of novel hits and their therapeutic targets, offering a way to obtain new therapies for unmet medical needs.
This project is carried out in collaboration with Dr. Manuel Collado (Instituto de Investigación Sanitaria de Santiago de Compostela, IDIS)
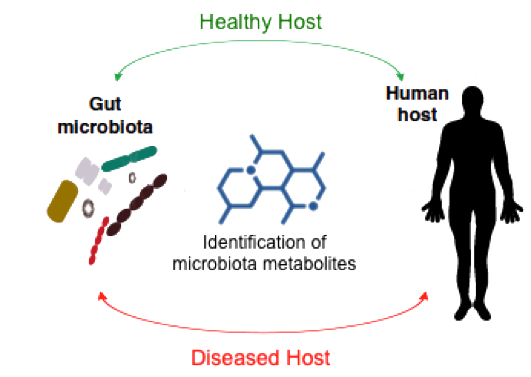
Identification of Microbiota Metabolites
The recent recognition that intestinal microbiota exerts profound effects on many aspects of human health and disease has led to the concept of a human meta-organism that integrates the communication between both prokaryotic and eukaryotic parts to achieve homeostasis. In particular, several diseases have been recently linked to the microbiota and their metabolites, including cancer, diabetes and obesity, cardiovascular disease, inflammatory bowel disease, autism, and immune system dysfunction. Furthermore, replacement of the microbiota of a diseased individual with that of a healthy individual clearly ameliorates certain diseases.
These findings strongly support the hypothesis that metabolites produced by the human microbiota regulate key biological functions in the host. However, little is known about the specific effectors that commensal bacteria use to interact with the human host because most of the microbiota metabolites are still to be discovered. In this line of research we are working on the development of a platform based on the use of labeled precursors and bioorthogonal chemistry to capture and identify specific microbiota metabolites. This knowledge will not only increase our understanding of their biological role but will also provide innovative pharmacological approaches.
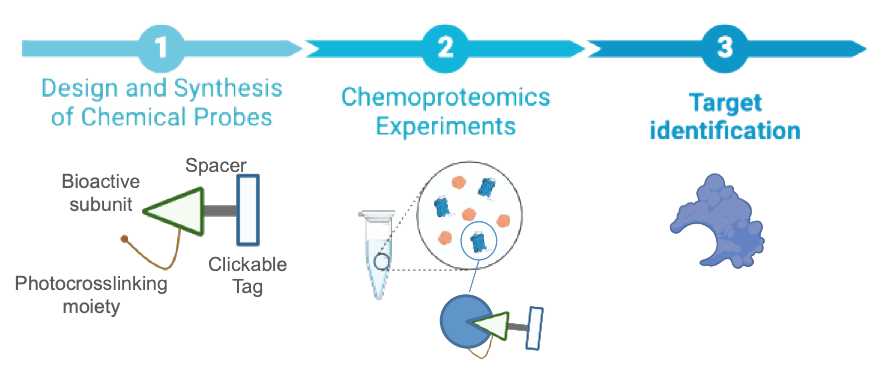
Chemical Probes for the Identification and Validation of Therapeutic Targets
Among the different approaches in drug discovery, identification of new therapeutic targets with the use of chemical probes stands out as one of the most powerful strategies for finding new mechanisms of action and for the development of new drugs. An area that could benefit most from such approach is the cannabinoid field. The variety of bioactive phytocannabinoids and synthetic compounds reported has led to the identification of a plethora of potentially beneficial therapeutic effects mediated by different molecular pathways. This makes a formidable challenge to ascribe a specific cannabinoid to a precise target. Among the cannabinoid ligands, cannabidiol (CBD) and WIN552122 are receiving a growing attention. CBD exerts important anti-inflammatory, antitumor and neuroprotective effects that can impact age-associated diseases, and it is the only cannabinoid approved for the treatment of severe forms of epilepsy. However, the biological actions of CBD itself are unclear and specific targets are mostly unknown. With respect to WIN552122, its recently reported immunomodulatory capacity could open new avenues for the development of novel vaccines for the prevention of allergy and other immune-mediated diseases. Hence, the development of chemical probes to profile the target landscape of CBD and WIN55212 is of utmost importance to associate specific proteins with observed therapeutic effects and eventually identify new drug targets for unmet medical needs.
Novel NMR Methods for the Study of Complex Biological Systems
Knowing the structure and behavior of molecules is crucial for understanding the world around us and for the development of new chemical products, drugs and materials. As our knowledge grows about how Nature works, the species under study steadily increase in size and complexity, making their analysis more and more difficult. Our research focuses on the development and application of cutting-edge NMR methods to reduce the time and effort needed to extract valuable chemical and biological information from complex systems, essential for the structural design of future drugs and biomaterials, and the identification of therapeutic targets.
Our key on-going research projects include:
• Development and application of advance solution-state NMR techniques.
• Biological studies by paramagnetic NMR.
• Characterization of the conformation and molecular recognition features of complex N-glycans.
• Determination of the structure, dynamics and binding properties of molecules with chemical and biological interest.
• Mixture analysis by NMR.
• NMR-based metabolomics.
• Pure shift NMR spectroscopy.
• 19F and 31P NMR spectroscopy.
• Diffusion NMR.
contact details for the nmr research line
Ángeles Canales Mayordomo: ma.canales@quim.ucm.es
Laura Castañar Acedo: lcastana@ucm.es